Con el fin de ganar flexibilidad, SOLID ha sido dividido en dos unidades funcionales y físicamente separadas: la unidad de preparación de muestras (SPU) para la recepción de la muestra, homogeneización, además de procesamiento, etc, y la unidad de análisis de la muestra (SAU), que alberga el microarray biosensor de anticuerpos en los ensayos inmunológicos. El diseño actual del instrumento es capaz de preparar 10 muestras, analizandolas en cinco matrices de anticuerpos individuales. Teniendo en cuenta este esquema de bloques, la distribución de masa es: SPU alrededor de 5,5 Kg y SAU menos de 2 kilogramos.
La Unidad de Preparación de la Muestra (SPU)
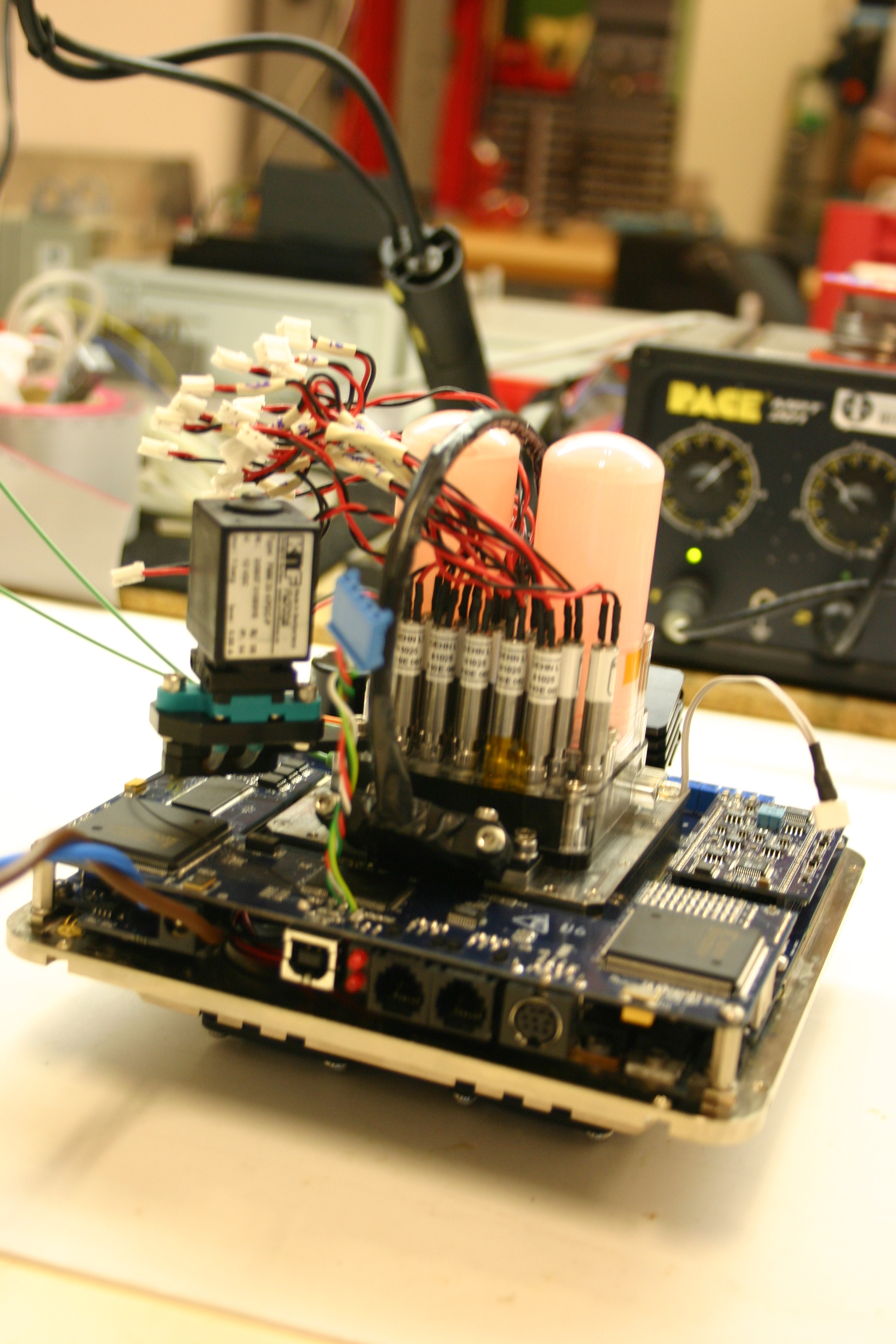
El prototipo de la SPU consiste en 10 celdas de extracción, cada una capaz de procesar de 0,1 a 1 g de material sólido y hasta 2 ml de buffer de extracción o otra muestra de líquido. La celda tiene un puerto de carga, que se sella después de que avance el extremo plano del sonicador. Para cada medición, se carga hasta 1 g de suelo, roca o suelo de hielo en la celda de extracción a través del puerto de carga. Entonces, la bocina del sonicador se mueve hacia adelante de modo que desplaza un anillo de cierre de membrana encerrando la muestra en una cámara hermética. El buffer de extracción se inyecta desde la dirección opuesta del flujo de salida, y despues se enciende el sonicador para realizar de 3 a 10 ciclos de tratamiento con ultrasonidos (1 min cada uno). La bocina de ultrasonidos junto con el anillo de membrana actúan como un pistón de empuja hacia adelante la muestra, la cual es filtrada a través de un filtro de tamaño de poro de 5-15 micras. La muestra filtrada puede ser entonces inyectada directamente a la cámara de microarrays de la SAU, donde será analizada por un inmunoensayo sándwich de microarrays (SMI) o por un inmunoensayo competitivo. Después de haber analizado la primera muestra, los tubos y válvulas SPU se enjuagan para evitar la contaminación cruzada entre las muestras y la obstrucción de las válvulas de material particulado.
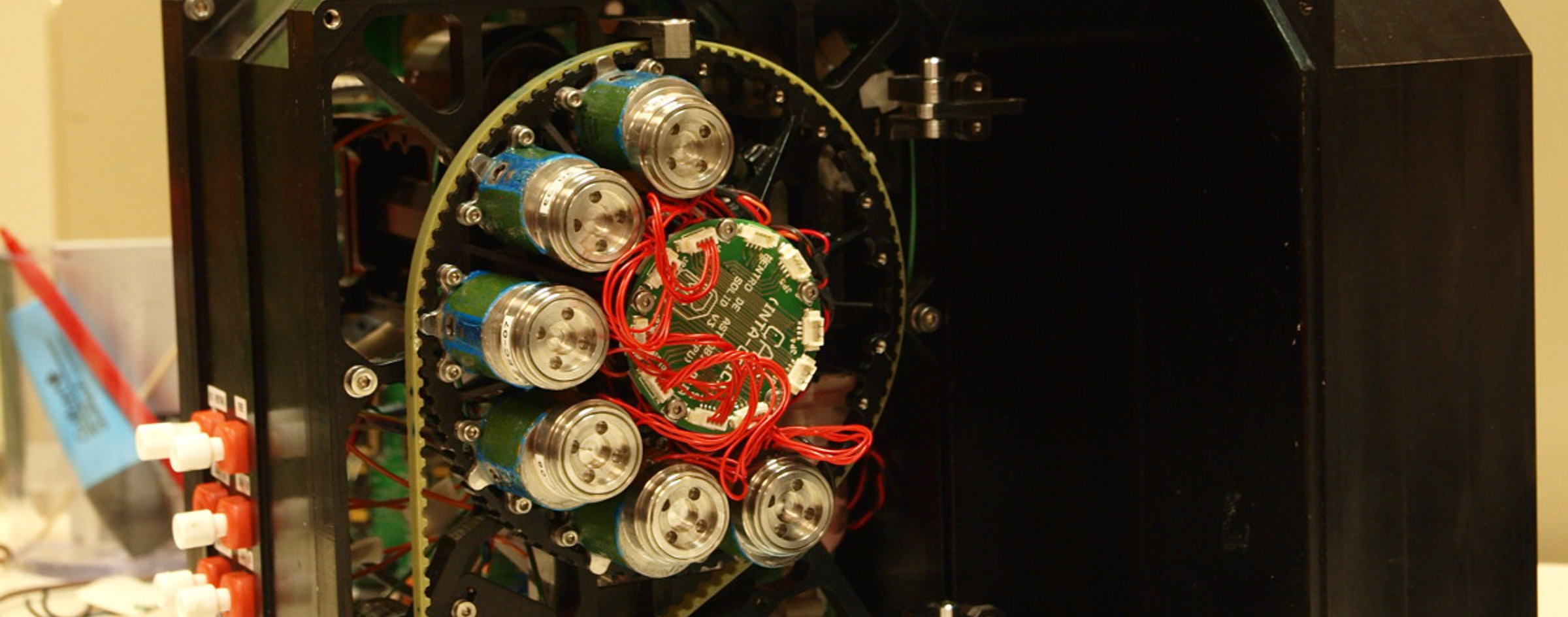
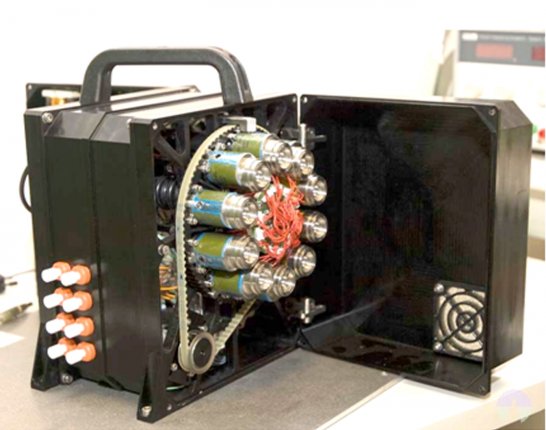
Instrumento SOLID3. (left) Unidad de Preparación de la Muestra (SPU) mostrando las 10 diferentes celdas de extracción para muestras de 0.5-1.0 g. (right) Unidad de Análisis de la Muestra (SAU) mostrando electrónica, bomba, depósitos de líquidos sobre el módulo de fluídica con cinco cámara de análisis, una por cada microarray.
La Unidad de Análisis de la Muestra (SAU)
La configuración actual de la SAU consta de un módulo de análisis, equipado con 5 céldas de flujo, que abarcan un microarray de anticuerpos cada una, así como la bomba, el manifold de fluídica, las válvulas, etc, necesarios para realizar el ensayo. La solución que contiene el analito de la SPU se incuba con el microarray de anticuerpos en la celda de flujo para llevar a cabo un inmunoensayo de sándwich, o un inmunoensayo competitivo. En ambos casos, después de que se complete el ensayo, el módulo de lectura de la señal se activa: un haz de láser entra por el borde frontal del soporte de microarrays y se transmite por él mediante reflexión total interna utilizando el mismo soporte como una guía de ondas. La luz excita los fluorocromos sobre las manchas y la señal de fluorescencia es capturada a través de una matriz de micro-lentes por un dispositivo CCD. Debido a que el instrumento fue concebido inicialmente para la exploración espacial, hemos diseñado un paquete óptico específico con el fin de capturar un área de superficie relativamente grande con la masa admisible mínima. Además, el paquete óptico compacto también permite una SAU compacta y ligera.
Antes de el análisis de la muestra, se requieren dos pruebas:
- En las pruebas para el proceso de captura de imagen, se toma una fotografía de todos los microarrays con dos objetivos: i) para comprobar el sistema de captura de imágenes (láser y CCD), y ii) para estimar el fondo óptico de la imagen.
- Para llevar a cabo un control negativo (en blanco) mediante la medición de la contribución de los anticuerpos fluorescentes en el fondo de la imagen. Para ello uno de los microarrays se incuba sólo con buffer (sin muestra) y se revela con anticuerpos fluorescentes. La imagen así obtenida se utiliza como una línea de base para eliminar todas las señales fluorescentes inespecíficas.
Debido al diseño modular de SOLID, la SAU puede consistir en cualquier número de celdas de flujo, y esto puede ser ajustado de acuerdo a las limitaciones de carga útil y objetivos de la ciencia. Hemos diseñado el instrumento de análisis con 5 células de flujo, para que sea capaz de llevar a cabo dos tipos de análisis de microarrays (inmunoensayo sándwich e inmunoensayo competitivo). Con este diseño, después de que se haya analizado la muestra, las celdas se pueden lavar con buffer para eliminar cualquier traza de la muestra original, y de ese modo la celda se puede cargar con una nueva muestra.
CÓMO FUNCIONA SOLID
Como tiene un diseño modular SOLID3 puede realizar 2 tipos diferentes de análisis dentro del mismo módulo SAU: un inmunoensayo de microarrays Sandwich (SMI), y un inmunoensayo de microarray competitivo (CMI). Se puede cambiar de un tipo a otro en función de los objetivos científicos y el funcionamiento del instrumento: CMI para detectar biomarcadores moleculares de vida extintas y otras moléculas pequeñas, y SMI para detectar complejos moleculares de vida existentes (proteínas, EPS, ácidos nucleicos, células enteras …).
Inmunoensayo de microarray Sandwich (SMI) en SOLID3.
Después de que la muestra se haya filtrado, sale de la SPU y se inyecta directamente a una celda en la SAU. Aquí, la muestra filtrada inunda una de las celdas de flujo y contacta con el microarray de anticuerpos. Un circuito de recirculación interna permite que la muestra se recircule durante un máximo de 1 h, para acelerar la cinética de la reacción entre antígenos y anticuerpos. Después del tiempo de incubación, la muestra sobrante se desecha al depósito de residuos y la celda de microarrays se lava con buffer de incubación para eliminar la muestra que no se ha unido. A continuación, se inyecta buffer en las cámaras auxiliares llenas de anticuerpos fluorescentes liofilizados, que los disuelven y que inundan la cámara de microarrays. Comienza entonces otra recirculación de hasta 1h. Para finalizar se produce un lavado adicional para eliminar el exceso de anticuerpos fluorescentes y se deja el microarray listo para la detección fluorescente. Un haz de laser se propaga mediante reflexión total interna (TIRF) a traves del soporte de vidrio que actúa como una guía de ondas. La señal fluorescente es capturada por un dispositivo CCD de alta sensibilidad que almacena una imagen .fits que puede ser procesada y analizada por un software convencional de microarrays. Como en el control negativo (en blanco), uno de los microarrays se incubaró sólo con buffer y se reveló con anticuerpos fluorescentes, la imagen obtenida en esa celda se utiliza como una línea base para eliminar todas las señales fluorescentes inespecíficas.
Esquema para un inmunoensayo sandwich en microarray.
Una señal positiva en un SMI indica:
i) La detección de una molécula con la misma estructura o muy parecida a la utilizada para producir el anticuerpo, y
ii) Esta molécula es parte de un complejo más grande, ya sea como un componente estructural de otra molécula o polímero, o simplemente capturado por macromoléculas amorfas, agregados o incluso partículas minerales.
Sea cual sea el resultado puede ser considerado como un signo directo o indirecto de vida el cual dependerá del tipo y número de diferentes moléculas detectadas. Una sóla señal positiva no puede ser tomada como prueba final para la presencia de moléculas biológicas, pero varias señales positivas con especificidades solapadas en varios anticuerpos representaría una evidencia clara de vida.
Inmunoensayo de microarray Competitivo (CMI).
Este formato de inmunoensayo es necesario para la detección de moléculas de pequeño tamaño tales como, por ejemplo, los biomarcadores moleculares para la vida extinta (hopanos y derivados, carotanes, etc) y, en general, todas las moléculas con un solo sitio (epítopo) para unirse al anticuerpo. Para el CMI, lo que se suele imprimir en el microarray son: antígenos, conjugados o moléculas target a 0,5 mg/ml como sondas de captura inmovilizadas. Anticuerpos fluorescentes previamente titulados se utilizan como trazadores, y antígenos, conjugados o moléculas target como competidores. Las curvas de calibración deben ser obtenidas previamente en el laboratorio como sigue: las diluciones en serie de moléculas target se incuban con su correspondiente anticuerpo fluorescente o con una mezcla de anticuerpos en un volumen total de 1 ml de buffer PBST – B (1xPBS, 0,01% de Tween20, 1% de BSA) durante 10 minutos. A continuación, se inyectan de forma manual en una cámara de la SAU ya inundada y se programa una recirculación de 10 minutos a 0,15 ml min -1 antes del lavado con 2 ml de buffer PBST – B durante 5 min a 0,4 ml min -1. El subsistema óptico se enciende y la imagen es capturada. Para el control en blanco, es decir, el ensayo que da la intensidad de la señal 100 % en cada punto, se sigue el mismo procedimiento pero sin antígenos o competidores en una cámara en paralelo. Las imágenes se procesan, y se determinan las intensidades de los microarrays con un software de microarrays. En los ensayos reales con SOLID, el CMI se hace pasando la muestra filtrada por la SPU a través de la cámara de anticuerpo fluorescente liofilizados de la SAU y se ponen a recircular durante 10 min sin pasar por el microarray, después se incuban en la cámara de microarrays durante otros 10 min. Y por último, después del lavado, se captura una imagen de CCD.
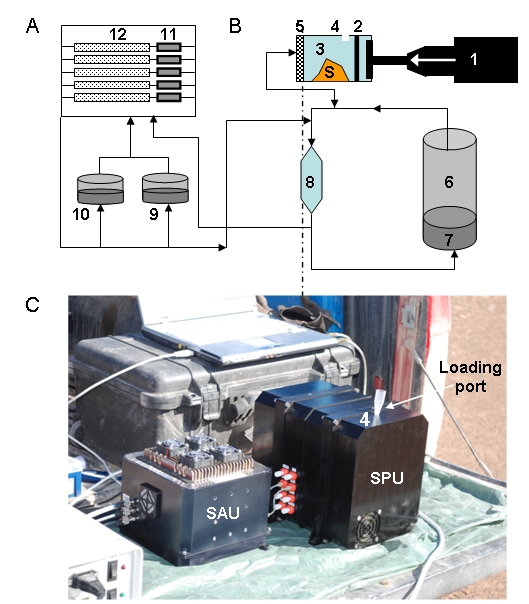
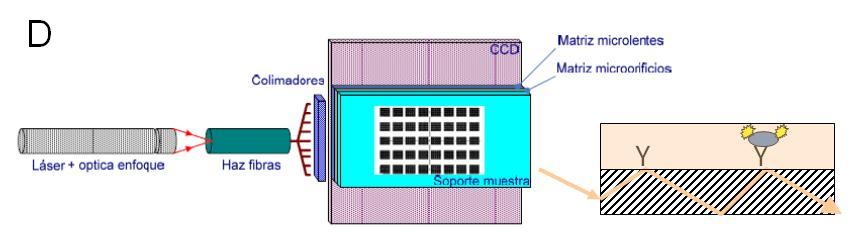
Diagrama funcional tanto de la SPU (A) como de la SAU (B). Una vez que una muestra (S) se ha cargado a través del puerto de muestreo (4), el aparato de ultrasonidos (1) encaja en la cámara de extracción (3) que arrastra hacia delante la membrana (2), dejando atrás el orificio de muestreo. La muestra es comprimida y herméticamente cerrada en un volumen más pequeño, antes de la adición de 2 ml de buffer de extracción desde el depósito (6) en dirección opuesta a través del filtro (5). El sonicador se activa para hacer varios ciclos de sonicación. Finalizada la sonicación, la bocina del sonicador se mueve hacia adelante forzando a la muestra a pasar a través del filtro. La muestra filtrada se queda en la llamada cámara de recirculación (8) y cuando sale de la SPU es para inundar las células de flujo de microarrays (12). Tanto la SPU y SAU tienen bombas y un conjunto de válvulas que permiten que la muestra llegue al lugar adecuado, y también permiten la recirculación a través de diferentes circuitos. (C) El módulo de lectura consiste en: un láser, óptica de enfoque, una fibra óptica para iluminar la parte delantera lateral del soporte de microarrays, el cual actúa como guía de onda para propagar la luz y excitar los fluorocromos, y un paquete óptico que comprende un array de pin-holes espaciados junto con una matriz de micro lentes, y un sensor CCD. (D) Esquema que muestra cómo la luz (flecha) se propaga a través de una guía de ondas (rayada) y excita a los fluorocromos retenidos por los anticuerpos (Y) a la derecha en la superficie
Hemos reportado recientemente la producción de un anticuerpo a ácido melítico y el desarrollo de un inmunoensayo de inhibición de microarrays fluorescente (IMI) para detectar esta sustancia a un límite de 5 ppb (ng ml -1). Se utilizó el anticuerpo anti-melítico para detectar ácido melítico en muestras de núcleos de perforación obtenidos de diferentes profundidades en el desierto de Atacama (Chile), un análogo terrestre de gran relevancia para Marte.
SENSIBILIDAD
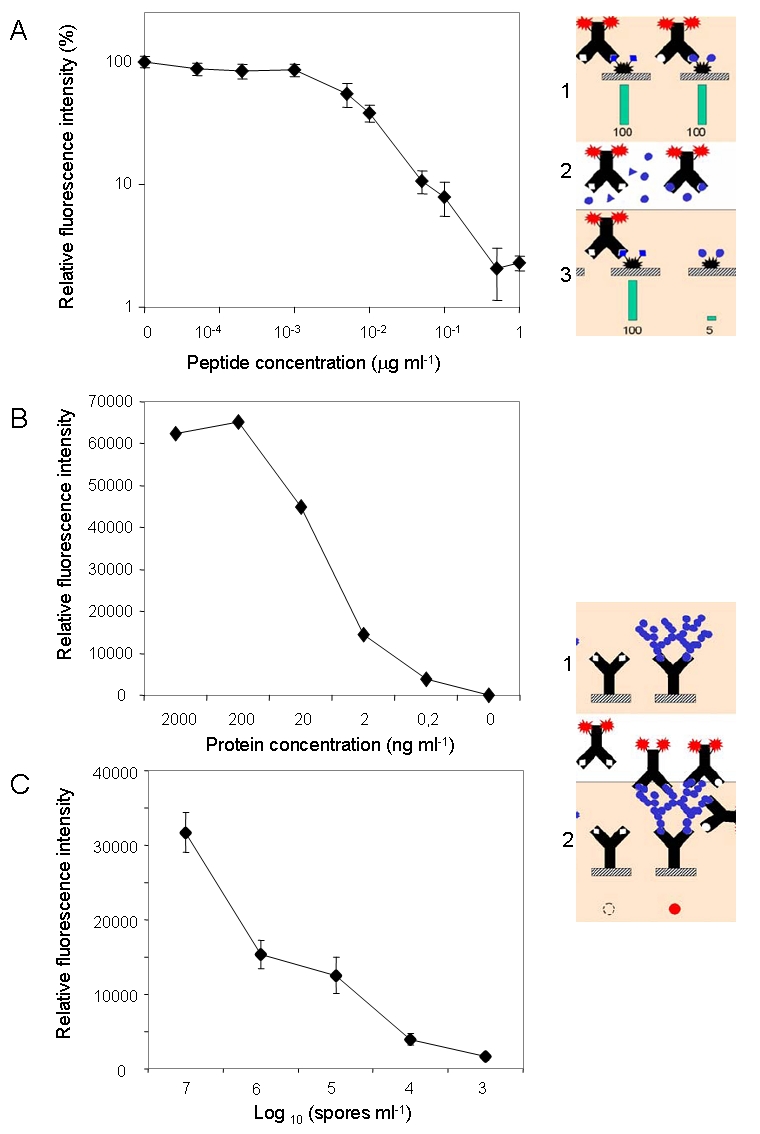
Hemos demostrado el rendimiento de SOLID para la detección de una amplia gama de compuestos de tamaño molecular, a partir del tamaño de aminoácidos, péptidos, proteínas, células enteras y esporas, con sensibilidades en 1-2 ppb (ng ml-1) de biomoléculas y 10 4-5 células ml -1 (Rivas et al., 2008; Parro et al., 2011). La sensibilidad es más dependiente de la calidad del anticuerpo que de la misma la instrumentación.
Determinación de la sensibilidad de los diferentes inmunoensayos con Solid 3: competitivo (A) y sandwich (B, C).
CAMPAÑAS
Tanto, el LDCHIP (Life Detector Chip) como SOLID han sido probados en el laboratorio y en varias campañas de campo. El trabajo con el prototipo SOLID2 fue especialmente relevante durante la campaña de 2005 del proyecto MARTE (Marte Technology Research analógica Experiment) en Río Tinto (Stoker et al., 2008, Parro et al., 2008; Parro et al., 2011). El LDChip300 y SOLID3 fueron probados durante la “AtacaMars2009” campaña de campo para el Desierto de Atacama en julio de 2009. El LDChip300 detectó compuestos biológicos de alto peso molecular en 0,5g de 2m de profundidad de perforación de núcleo. (Parro et al., 2011; Fernández-Remolar et al., 2013). Del mismo modo, SOLID3 fue probado en la isla Decepción (Antártida) en dos campañas de campo (2010 y 2012) con permafrost, muestras superficiales (suelos y rocas), (Blanco et al., 2012), y muestras hidrotermales. Por último, una nueva versión de SPU que contiene una sola celda de extracción reutilizable, ha sido probado recientemente en una campaña de campo para el Ártico canadiense, en particular a la Estación de Investigación del Ártico McGill (MARS). SOLID detectó microbios psychrophile en manantiales ricos sulfato y el permafrost del Ártico .
In order to gain flexibility, SOLID has been divided into two main functional and physically separate units: the sample preparation unit (SPU) for sample allocation, homogenization, further processing, etc., and the sample analysis unit (SAU), bearing the antibody microarray biosensor for the immunological assays. Current design of the instrument is able to run 10 samples by using five single arrays. Considering this block scheme, the mass distribution is: SPU around 5,5 Kg and SAU less than 2 Kg.
The Sample Preparation Unit (SPU)
The SPU prototype consists of 10 extraction cells, each one capable to process from 0.1 to 1 g of solid material and up to 2 ml of extraction buffer or another liquid sample. The cell has a loading port, which is sealed after the advance of a sonication horn. For each measurement, up to 1 g of soil, ground rock or ice is loaded into the extraction cell through the loading port. Then, the sonicator horn moves forward so that it displaces a close membrane-ring confining the sample in a hermetic chamber. The extraction buffer is injected from the opposite direction of the exit flow, and the ultrasonicator is powered on to perform from 3 to 10 cycles of ultrasonication (1 min each). The sonicator horn and membrane-ring acting as a piston push forward the sample, which is filtrated through 5-15 microns pore size filter. The filtrate can then be directly injected to the microarray chamber of the SAU, where it will be analyzed by a sandwich microarray immunoassay (SMI) or competitive immunoassay. After the first sample has been analyzed, the SPU pipes and valves are rinsed to avoid cross-contamination between samples and obstruction of the valves by particulate matter.
SOLID3 instrument. (left) Sample Preparation Unit (SPU) showing 10 different extraction cells for 0.5-1.0 g sample. (right) The Sample Analysis Unit (SAU) showing electronics, pump, liquid deposits over a module with 5 flow cells, each one for one microarray analysis.
The Sample Analysis Unit (SAU)
The current configuration of the SAU consists of one analysis module, equipped with 5 flow cells, which allocate one antibody microarrays each, as well as the motor, pumps, fluidics, valves, etc., necessary to perform the analysis. The analyte-containing solution from the SPU is incubated with the antibody microarray in the flow cell to perform a sandwich microarray immunoassay, or a competitive microarray immunoassay. In both cases, after the assay is completed, the Signal Readout Module subsystem is activated: a laser beam enters by the front edge of the microarray support and is transmitted by total internal reflexion using the same support as a waveguide. The light excites the fluorochromes on the spots and the fluorescence signal is captured through a micro-lens array by a CCD device. Because the instrument was initially conceived for space exploration, we designed a specific optical package in order to capture a relatively big surface area with the minimal mass allowable. In addition, the compact optical package also allows a compact and light SAU.
Two tests are required before the analysis of the sample:
- Testing for the image capturing process. A picture of all the microarrays is taken for two purposes: i) to check the image capturing system (laser and CCD), and ii) to estimate the optical background of the image.
- To conduct a blank control by measuring the contribution of the fluorescent antibodies to the background of the image. For this purpose one of the microarrays is incubated just with buffer (no sample) and revealed with fluorescent antibodies. The image thus obtained is used as a base line to eliminate all the unspecific fluorescent signals.
Due to the modular design of SOLID, the SAU can consist of any number of flow cells, and this can be adjusted according to payload limitations and science objectives. We have chosen an instrument design based on one analysis module with 5 flow cells, which is capable of conducting two types of analyses (sandwich microarray immunoassay and competitive microarray immunoassay). Under this design, after a sample has been analyzed, the cells can be washed with buffer solution once more to remove any traces of the original sample, and the cell can be loaded with a new sample.
HOW SOLID WORKS
Due to its modular design, SOLID3 can perform 2 different types of analysis within the same SAU module: a Sandwich microarray immunoassay (SMI), and a Competitive microarray immunoassay (CMI). It can switch from one type to the other depending on scientific goals and instrument performance: CMI to detect extinct life molecular biomarkers and other small molecules, and SMI to detect extant life molecular complexes (proteins, EPS, nucleic acids, whole cells…).
Sandwich microarray immunoassay (SMI) in SOLID3.
After the sample has been filtered, the filtrate leaves the SPU and is directly injected to cell1 in the SAU. Here, the filtrated sample floods one of the flow cells and contact the antibody microarrays. An internal recirculation circuit allows the sample to be recirculated for up to 1 h, in order speed reaction kinetics between antigens and spotted antibodies. After the incubation time, the remaining sample is discarded into the waste deposit and the microarray cell is washed out with incubation buffer to remove the non-bound sample. Then, buffer is injected to the auxiliary chambers filled with lyophilized fluorescent antibodies, let to dissolve, flood the microarray chamber, and set for recirculation for up to 1h. An additional wash remove the excess of fluorescent antibodies and leaves the microarray ready for fluorescent detection by excitation with a laser beam by total internal reflexion (TIRF) through the glass support which acts as a waveguide. The fluorescent signal is captured by a high sensitive CCD device and store a .fits image that can be processed and analyze by conventional microarray software. As a blank control, one of the microarrays are just incubated with buffer and revealed with fluorescent antibodies. The obtained image is used as a baseline to eliminate all the unspecific fluorescent signals.
Scheme for a sandwich microarray immunoassay.
A positive signal in an SMI indicates:
i) Detection of a molecule with the same or highly related structure to that used to produce the antibody, and
ii) This molecule is part of a bigger complex, either as a structural component of another molecule or polymer, or just captured by amorphous macromolecules, aggregates or even mineral particles.
Whether this result can be considered as a direct or indirect sign of life would depend on the type and number of different molecules detected. Just one positive may not be taken as a final proof for the presence of bio-molecules, but several positives with overlapping specificities in several antibodies would represent clear evidence of life.
Competitive microarray immnuno-assay (CMI).
This immunoassay format is necessary for the detection of small size molecules such as, for example, those molecular biomarkers for extinct life (hopanes and derivatives, carotanes, etc) and, in general, all molecules with just one site (epitope) to bind to the antibody. For the CMI, what it is usually printed in the microarray is: antigens, conjugates or target molecules at 0.5 mg/ml as immobilized capturing probes. Previously titrated fluorescent antibodies are used as tracers, and the antigens, conjugates or target molecules as competitors. Calibration curves must be previously obtained in the laboratory as follows: serial dilutions of target molecules are incubated with their corresponding fluorescent antibody or with an antibody mixture in a total volume of 1ml of PBST-B buffer (1 x PBS, 0.01% Tween 20, 1% BSA) for 10 minutes. Then, they are manually injected to an already flooded SOLID’s SAU microarray chamber and set to recirculate for 10 minutes at 0.15 ml min-1 before washing with 2 ml of PBST-B buffer for 5 min at 0.4 ml min-1. The optical subsystem is powered on and a CCD image is captured. For the blank control, that is, the assay that gives the 100% signal intensity in each spot, the same procedure is followed but without antigens or competitors in a parallel chamber. The images are processed, the spot intensities of the microarrays determined, and the data analyzed by microarray Software. For actual test samples, CMI is done by passing the filtrate from SPU through the lyophilized fluorescent antibody chamber and set to recirculate for 10 min before incubating for another 10 min to the cell 2 microarray chamber. After washing, an image is captured by CCD.
Functional diagram of both the SPU (A) and the SAU (B). Once a sample (S) has been loaded through the sampling port (4), the sonicator (1) fits into the extraction chamber (3) and moves forward the membrane (2), leaving behind the sampling hole. The sample in compressed and hermetically confined in a smaller volume before adding 2 ml of extraction buffer from deposit (6) in opposite direction through the filter (5). The sonicator is activated and several sonication cycles can be done. Then the sonicator horn moves forward to force the sample to pass through the filter. The filtrate then goes to the so called recirculation chamber (8) and then leaves the SPU to flood the microarray flow cells (12). Both the SPU and SAU have pumps and a set of valves that allow the sample to reach the appropriate place, and also allow recirculation through different circuits. (C) The signal read-out module consisting of: a laser, focus optics, a fiber optic to illuminate the side front of the microarray support, which acts as a waveguide to propagate light and excite the fluorochromes, and an optical package comprising a pin-holes array spacer, a micro lens array, and a CCD sensor. (D) Scheme showing how light (arrow) propagates through a waveguide (hatched) and excite the fluorochromes retained by the antibodies (Y) right on the surface
We have recently report the production of an antibody to mellitic acid and the development of a fluorescent inhibition microarray immunoassay (IMI) to detect this substance to a limit of 5 ppb (ng mL-1). We used the anti-mellitic antibody to detect mellitic acid in drill core samples obtained from different depths in the Atacama Desert (Chile), a highly relevant terrestrial analogue for Mars.
SENSITIVY
We have demonstrated the performance of SOLID for the detection of a broad range of molecular size compounds, from the amino acid size, peptides, proteins, to whole cells and spores, with sensitivities at 1-2 ppb (ng ml-1) for biomolecules and 104-5 cells ml-1 (Rivas et al., 2008; Parro et al., 2011). The sensitivity is more dependent from the quality of the antibody than the instrumentation itself.
Determining the sensitivity of different immunoassay with SOLID 3: competitive (A) and sandwich (B,C).
FIELD CAMPAIGNS
Both, LDCHIP (Life Detector Chip) and SOLID have been tested in the laboratory and during several field campaigns. Especially relevant was the work with SOLID2 prototype during the 2005 campaign of the MARTE (Mars Analogue Research Technology Experiment) project in Rio Tinto (Stoker et al., 2008, Parro et al., 2008; Parro et al., 2011). LDChip300 and SOLID3 were tested during the “AtacaMars2009” field campaign to the Atacama Desert in July 2009. The LDChip300 detected high molecular weight biological compounds in 0.5 g of a 2 m deep drill core (Parro et al., 2011; Fernández-Remolar et al., 2013). Similarly, SOLID3 was tested in Deception Island (Antarctica) in two field campaigns (2010 and 2012) with permafrost, superficial samples (soils and rocks) (Blanco et al., 2012), and hydrothermal samples. Finally, a new version of SPU containing a single, reusable, extraction cell has been recently tested in a field campaign to the Canadian Arctic, particularly to the McGill Arctic Research Station (MARS). The SOLID detected psychrophile microbes in sulfate rich springs and the Arctic permafrost.