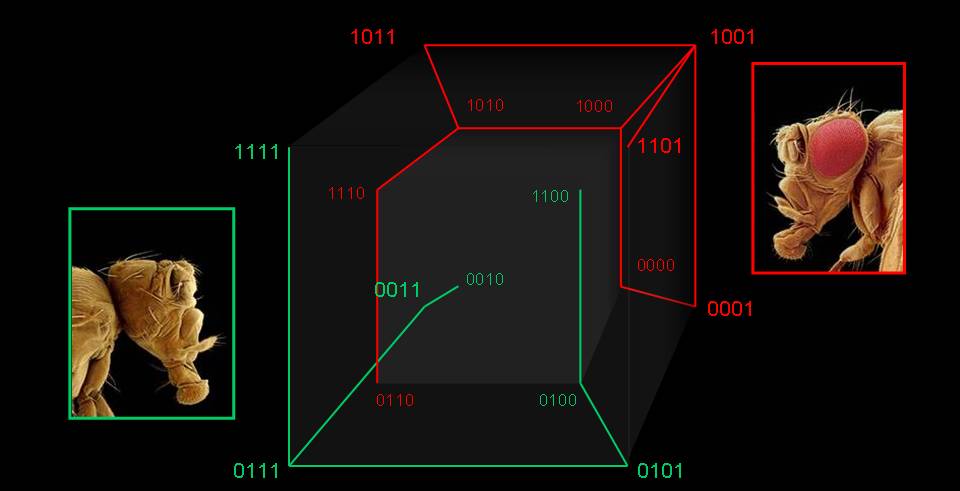
The redundancy between genotype and phenotype is present at many levels: when sequences map to structures, in networks of functional proteins, in regulatory networks, in metabolism, and way up to whole organisms. Actually, we cannot think of any evolutionarily meaningful genotype-phenotype (GP) map that does not present an extreme degeneration. Genotype networks occupy vast regions of genome spaces and allow a costless navigation of those spaces. Further, small changes in genotype occasionally cause huge changes in phenotype: an evolutionary innovation may be just a few mutations away. In this way, diversity can be maintained and, when necessary, innovative phenotypes may be found close to currently realized ones. Not only that, but exaptation appears to be extremely common in any realistic GP model. We have characterized the dynamics of populations of replicators on generic networks, and have shown that properties like network connectivity or network assortativity lead to non-trivial population dynamics, as an acceleration in the ticking rate of the molecular clock. The topological properties of genotype spaces induced by simple GP models might be universal. Despite the sustained growth of the area in the last decade, there is a multiplicity of open questions of the highest relevance to develop an updated evolutionary theory.
 RNA, THE ORIGIN OF LIFE AND MOLECULAR EVOLUTION
RNA, THE ORIGIN OF LIFE AND MOLECULAR EVOLUTION
RNA is a very versatile molecule. It may codify genetic information, as in extant RNA viruses, and can perform catalytic activity, as proteins do. This double role supports the plausibility of a prebiotic RNA world. We use RNA sequences and their folded, minimum free energy configuration, as a working example of the genotype-phenotype map. Among the questions we approach by means of this model are the adaptation of heterogeneous populations (where mutations act at the genotypic level but natural selection affects the phenotype) or the emergence of chemical function in the absence of template replication. We have shown that the extreme redundancy of the sequence-structure map in RNA may be behind the appearance of complex molecules by means of modular evolution. Exhaustive folding of the sequence space of short RNAs (up to 20 nucleotides) is giving us important clues on the topological structure of generic genotype-phenotype maps, and on the characteristics of models of phenotypic innovation. Theoretical and computational studies with circular RNA molecules have provided reliable support to a de novo emergence of viroid-like replicons.
 VIRAL EVOLUTION AND ANTIVIRAL THERAPIES
VIRAL EVOLUTION AND ANTIVIRAL THERAPIES
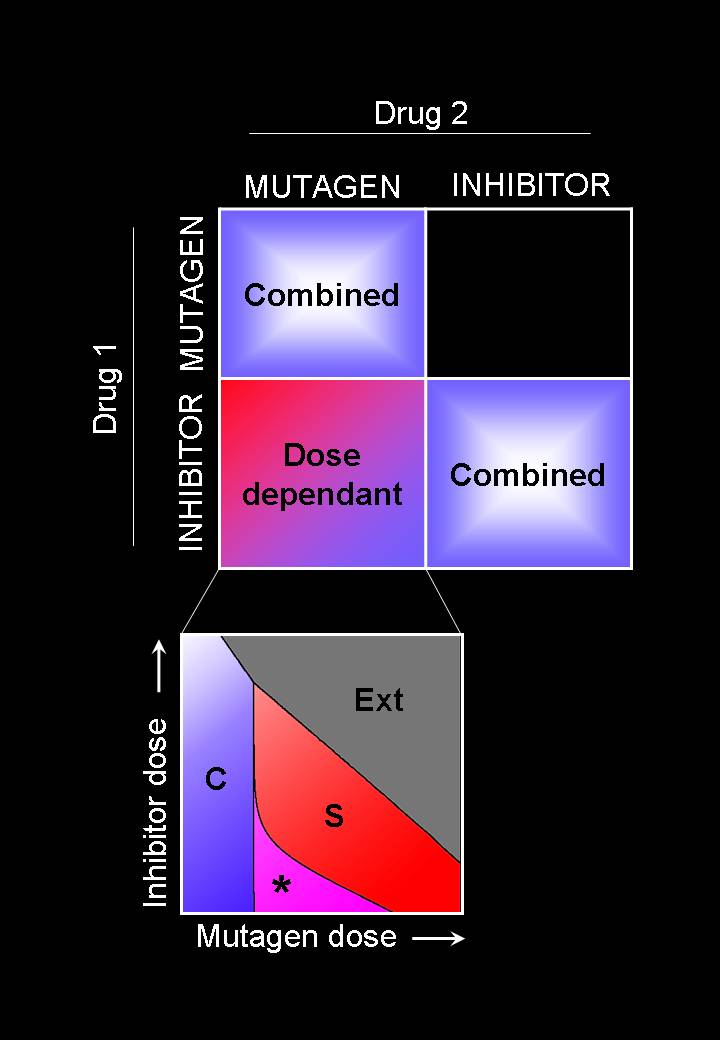
Viruses are amongst the “simplest” replicators. We are especially interested in the dynamics of RNA viruses and in the molecular mechanisms that generate their diversity and permit their fast adaptation. Among others, we have investigated the role of generic mechanisms such as variable mutation rates or the effect of genome fragmentation. For more than a decade by now, we have analysed and modelled experimental results obtained by our collaborators with a number of different viruses and phages (vesicular stomatitis virus, foot-and-mouth disease virus, beta-phage, and recently hepatitis C). These viruses share similarities that we try to capture in generic models of viral dynamics, but also present important differences that condition, for instance, the success of antivial therapies. We have introduced a model of viral adaptation able to predict in controlled conditions the response of the virus to the action of dissimilar antiviral drugs. Among our current interests stars the evolutionary origin of multipartite viruses and satellite viruses, and the role space may have played in their emergence and persistence.
 LINGUISTIC DIVERSITY, CULTURAL EVOLUTION
LINGUISTIC DIVERSITY, CULTURAL EVOLUTION
Ecology and culture share an amazing number of similarities. Species and languages reach their maximum diversity at the equator, present similar species-area relationships, and their abundances (number of speakers of a language or number of individuals in a species) more than often follow similar distributions. We have studied linguistic diversity is space by using the most comprehensive database available, the Ethnologue. We have identified an allometric relationship between the area occupied by the speakers of a language (often assimilated to a “culture”) and the population size, and have hypothesized that demographic dynamics and unavoidable conflicts between neighbouring groups explain the pattern. By analysing the correlation between linguistic diversity at the local scale and a large number of environmental variables, we have shown that river density and landscape roughness play a determinant role in regions rich in languages. A systematic study of the properties of networks of spatial contacts between linguistic groups has revealed that language contact networks have low intervality. A model of an adaptive network based on demography, conflicts and effective interactions between neighboring cultures reproduces all patters uncovered. In the broader context of cultural evolution, we have shown that epistatic interactions among cultural traits might be behind a class of cultural paradigm changes.